Prostate cancer patients who do not respond well to PSMA-targeted therapy often have potentially treatable mutations in their DNA damage-repair genes, according to research published in the May issue of The Journal of Nuclear Medicine. In an exploratory study using a relatively new technique—targeted next-generation gene sequencing—researchers found mutations in six out of seven patients with metastatic castration-resistant prostate cancer who did not respond to prostate-specific membrane antigen (PSMA)-targeted radiopharmaceutical therapy despite sufficient PSMA expression in their tumors.
Prostate-specific membrane antigen-targeting α-radiation therapy (PSMA-TAT) with 225Ac-PSMA-617 is an emerging and promising approach to treating metastatic castration-resistant prostate cancer. However, a reported 37 percent of patients respond poorly to this therapy even though they have sufficient PSMA expression. Preclinical and early studies suggest that particular DNA damage-repair-associated gene mutations can either increase or decrease the radiosensitivity of prostate cancers and may influence a patient’s response to radiopharmaceutical therapy.
“Given the important role of DNA damage-repair gene alterations, we sought to evaluate the frequency of these defects in patients with an insufficient response to 225Ac-PSMA-617 therapy with next-generation gene sequencing,” said Clemens Kratochwil, MD, supervising physician for radionuclide therapy at Heidelberg University Hospital in Heidelberg, Germany. “A relatively new technique, targeted next-generation sequencing, allows rapid analysis of an individual tumor’s genome. Theoretically, such information can be predictive as to whether a patient has an increased probability to benefit from one specific treatment or combination therapy.”
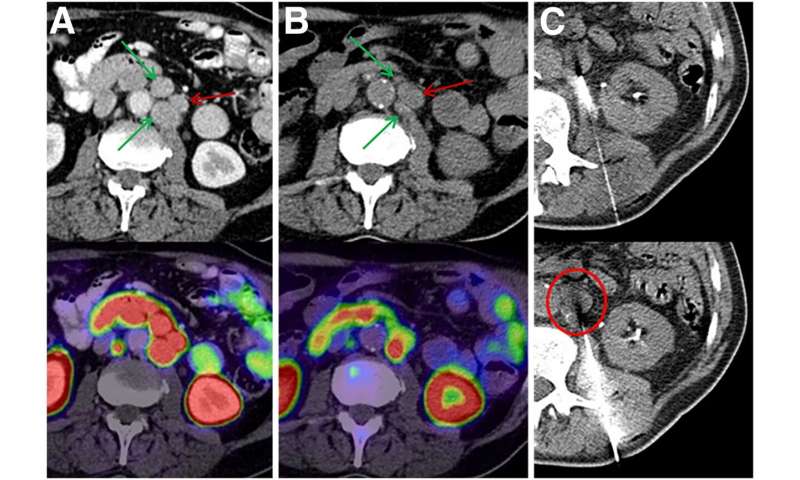
From a group of 60 patients treated with 225Ac-PSMA-617 PSMA-TAT, researchers identified 10 patients with a poor therapy response despite sufficient PSMA expression. CT-guided biopsies were obtained from seven of the 10 patients and then validated with histopathology and immunostaining. Biopsy specimens were analyzed with two separate targeted next-generation-sequencing panels to detect mutations in 37 genes involved in DNA-damage recognition, checkpoint signaling or DNA-damage repair.
In the seven biopsy specimens, researchers found a total of 15 whole-gene deletions, deleterious mutations and presumably deleterious mutations. Loss of function alterations were found more than once in TP53, ATM and CHEK2, with one patient even having a co-mutation of TP53 and CHEK2. Further loss-of-function mutations or whole-gene deletions were detected once in the analyzed sample set for BRCA1, BRCA2, PMS1, NBN, MSH2, MSH6, PALB2 and FANCB, respectively. For some other alterations found in ATM, BRCA1, MSH2, SLX4, ERCC and various FANC genes, it was not clear whether they negatively affected its function or were non-pathological variants as part of the normal human diversity.
“Whether the efforts and costs of next-generation sequencing are warranted in a dedicated clinical situation will depend on several factors,” noted Kratochwil. “First, the mutation must be druggable in order to be led to a change in therapeutic management. Second, if the mutations are very rare, then sophisticated testing would not be cost- or time-effective. Lastly, if many patients were found to have comparable mutations, individual testing could be avoided and a one-size-fits-all approach could be applied. The significance of this preliminary work is in guiding of further research, and our findings will likely lead to a change of research practice in the near future.”
Source: Read Full Article
